This package is geared towards the calculation of GCI, a novel CGM metric aimed at capturing the shape of glucose variability using the periodogram decomposition, in two distinct pathways. For the purposes of this document, we first generate a random time series meant to behave like a CGM sensor signal with corresponding time stamps.
set.seed(1234)
# Two weeks of data, sampled every 15 minutes
cgm_times = seq(from=as.POSIXct("2000-01-01"),
to = as.POSIXct("2000-01-01") + as.difftime(17, units = "days"),
by=15*60)[1:1344]
# Simple AR(1) with coefficient 0.5 used for example purposes
cgm_data = arima.sim(list(ar=.5), n=1632)[1:1344]Step-by-Step Procedure
For the first procedure, the user is able to manually perform all 3 requisite steps towards calculation of GCI: estimation of the log-spectral density through the smoothed log-periodogram, summarization of the variability decomposition using disjoint piece-wise linear models, and combination of the resulting summary into a weighted linear combination to calculate GCI.
To calculate the estimate of the log-spectral density, the user need only provide the CGM data and corresponding timestamps to the “pgram()” function, which uses a set of reasonable defaults for trimming, filtering, and smoothing parameters (all of which can be overwritten through the “options” argument). This allows for examination of the smoothed log-periodogram itself.
lpgram = pgram(cgm_data, cgm_times)The resulting smoothed log-periodogram can then be visualized in the following fashion.
plot(lpgram$freqs, lpgram$lpgram, type = "l",
main = "Smoothed log-periodogram of AR(1)",
xlab = "Frequency", ylab = "Log-Spectral Density Est.")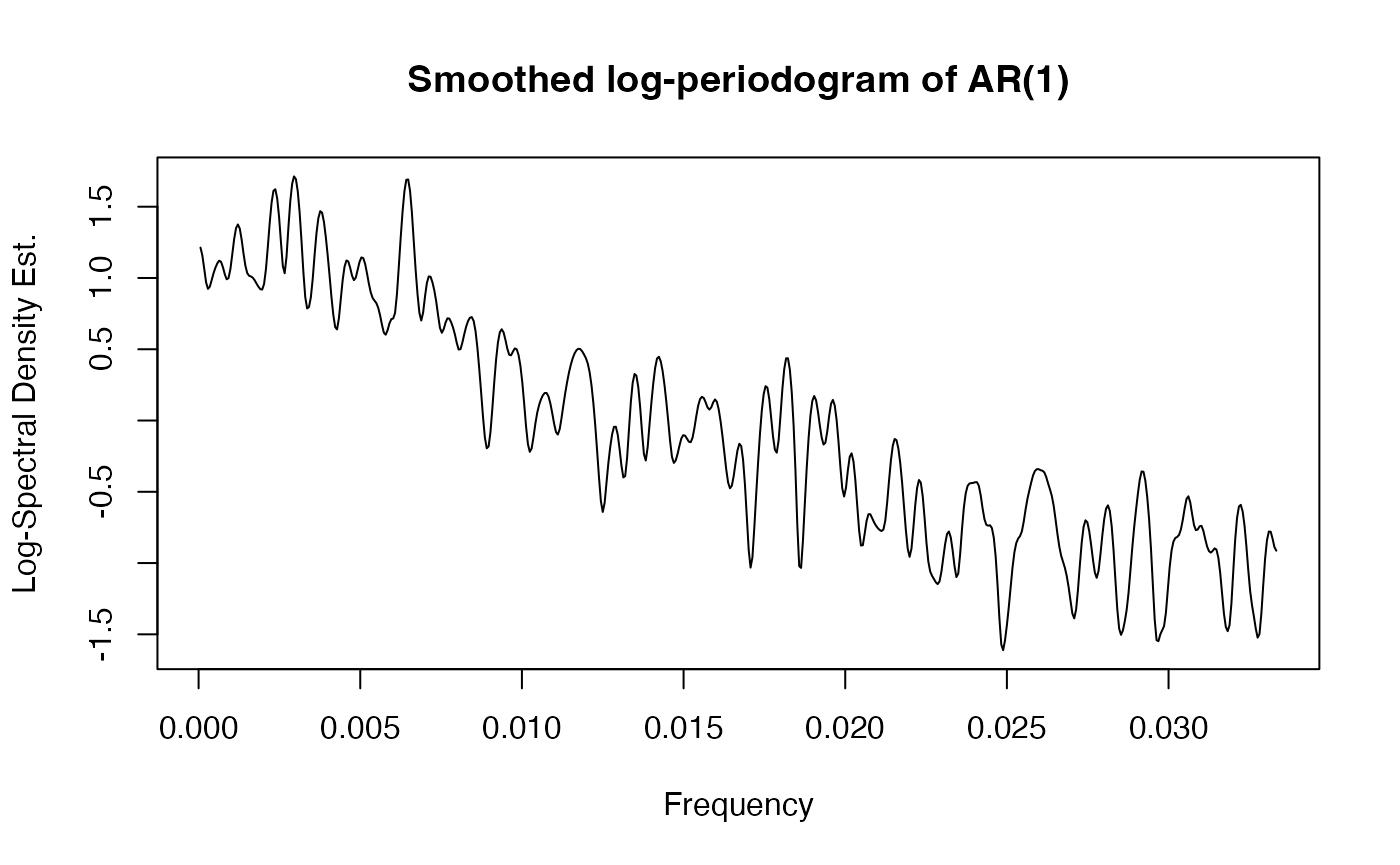
The user can then use the object output by “pgram()” to calculate the 6 degree of freedom disjoint linear model summary of the smoothed log-periodogram. This choice for summarization of periodogram structure relates to features observed in exploratory data analyses upon the HYPNOS study population. Each of these metrics indicate an aspect of glucose variability (the full group being summarized by the GCI), making them viable metrics in and of themselves. It is for this reason that these values are made available through this procedure.
sum6DF = linear_approx(lpgram)Finally, the user can calculate the GCI as the weighted linear combination of the disjoint linear model parameters, where the weightings were determined by canonical correlation analysis (CCA) upon the HYPNOS population. This technique was used to find the linear combination of the linear model parameters which was most consistent over the two HYPNOS wear periods. This weighting is provided as the default “weights” argument to “calculate_GCI()”.
seq_GCI = calculate_GCI(sum6DF)Simplified Procedure
For users which do not need or desire access to intermediate objects (the estimate of the log-spectral density and the 6 degree of freedom linear model parameters), rGCI allows for passing a dataframe with the CGM data in long format to the “calculate_GCI()” function, as opposed to an object returned by “linear_approx()”. This dataframe must contain the CGM sensor readings in a column named “glucose” and the corresponding timestamps in a column named “timestamp”.
df_GCI = calculate_GCI(data.frame(glucose = cgm_data, timestamp = cgm_times))